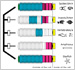
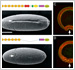
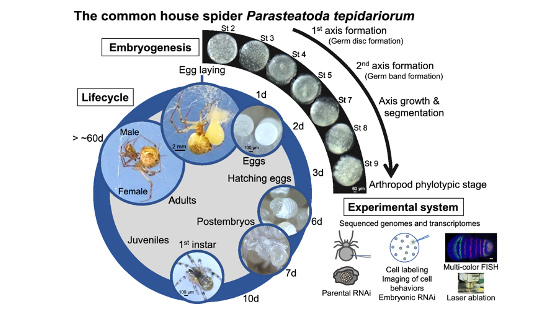
To understand the deep origins of animal diversity, we study mechanisms of development and cell-cell adhesion mainly using Drosophila and the common house spider Parasteatoda tepidariorum.
Go to Japanese.
News
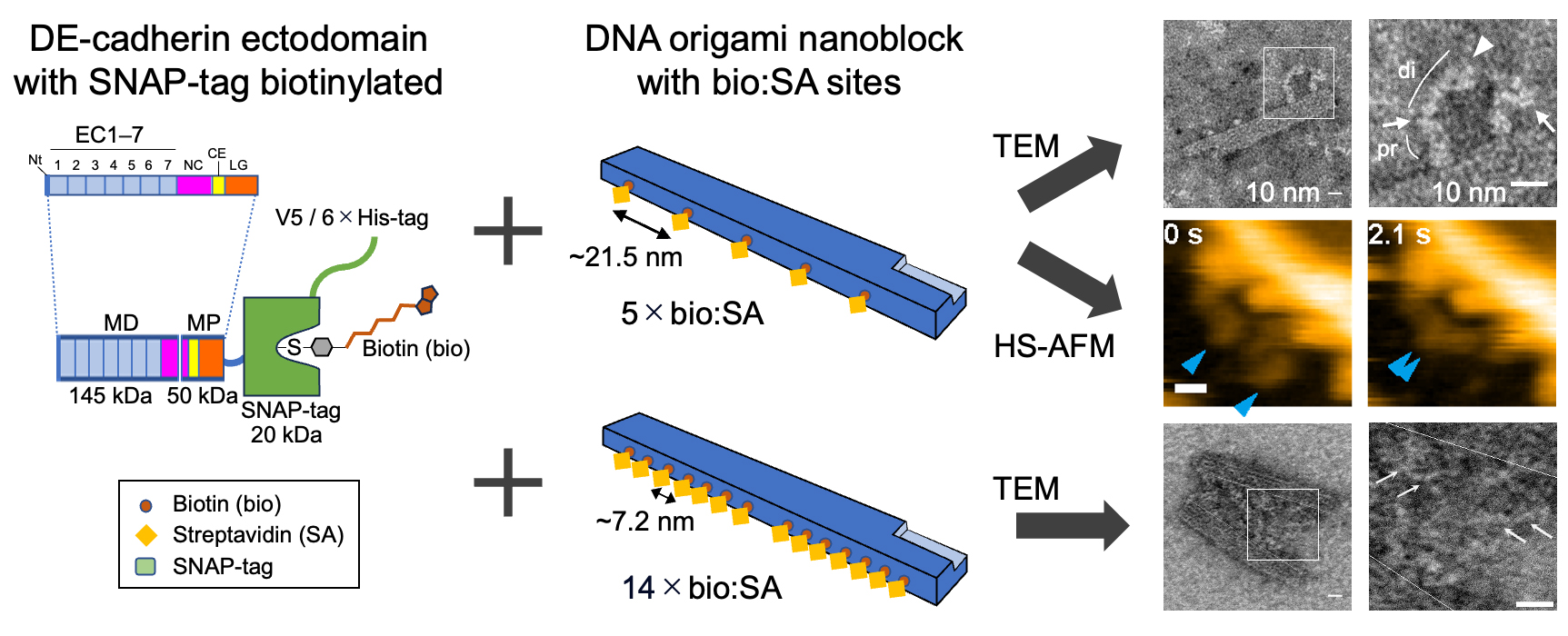
24.12.09 Our paper has been published in Journal of Molecular Biology: Nanoscale Visualization of Drosophila E-cadherin Ectodomain Fragments and Their Interactions Using DNA Origami Nanoblocks.
24.08.27 Students from University Giessen learned spider experimental techniques in our lab for three days. See here
23.04.01 Ren Negishi, a new student, has started to work in our laboratory.
23.03.23 Yasuko gave a talk about mechanisms of pattern formation in early spider embryos at Princeton University.
22.11.09 Yasuko gave a talk about her ongoing work on single-cell/single-nuc RNA-seq analyses of spider embryos at Franco-Japanese Developmental Biology Meeting held in Strasbourg, France.
22.10.06 ![]() Iwasaki and Nanjo's paper was published in BMC Biology. "Lineage‑specific, fast‑evolving GATA‑like gene regulates zygotic gene activation to promote endoderm specification and pattern formation in the Theridiidae spider."
Iwasaki and Nanjo's paper was published in BMC Biology. "Lineage‑specific, fast‑evolving GATA‑like gene regulates zygotic gene activation to promote endoderm specification and pattern formation in the Theridiidae spider."
22.0812 Fujiwara's paper was published today in Frontiers in Cell and Developmental Biology: "Virtual spherical-shaped multicellular platform for simulating the morphogenetic processes of spider-like body axis formation."
22.07.22 Yasuko's paper was published today in Frontiers in Cell and Developmental Biology: "Reconstruction of the global polarity of an early spider embryo by single-cell and single-nucleus transcriptome analysis."
21.10.21 Hiroki Oda was invited to give a talk at Virtual Gastrulation Zoom Talks (VGZT). "Axis formation in spider embryos: Regulative development under the microscope"
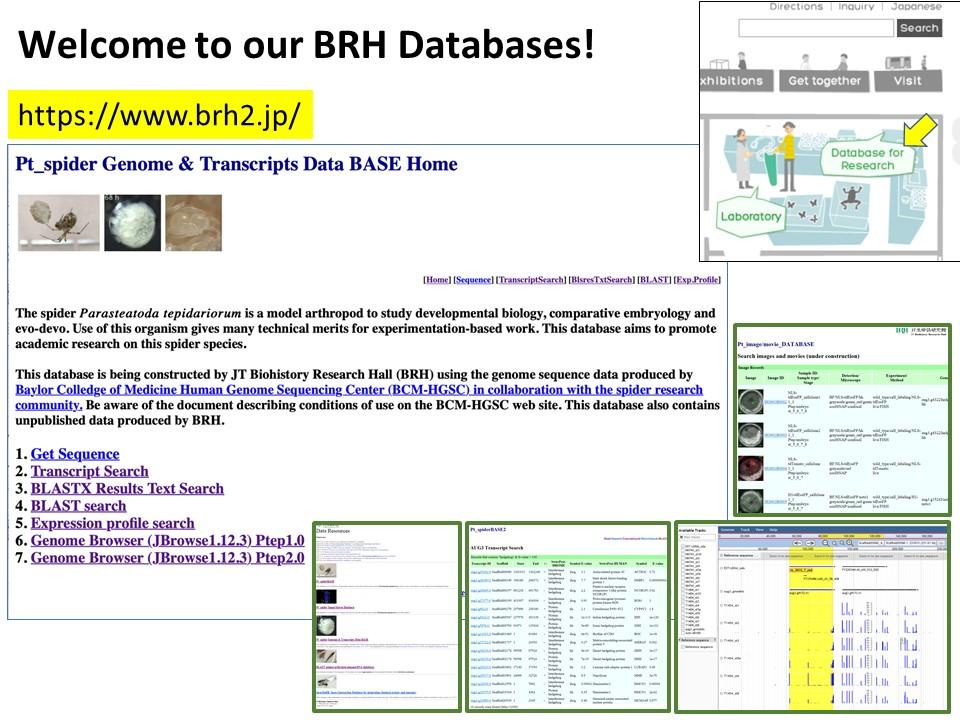
2021.9.30 The spider Parasteatoda tepidariorum Genome Database has been updated to version 3.
21.07.29 An interview with the first author Dr. Shigetaka Nishiguchi appeared in Journal of Cell Science.
21.07.20 Lecture slides used by Hiroki Oda in ISP2021 Osaka Univ are available from this link.
21.06.22 ![]() Our study of visualizing cadherin ectodomains by atomic force microscopy has been published in Journal of Cell Science.
Our study of visualizing cadherin ectodomains by atomic force microscopy has been published in Journal of Cell Science.
21.02.20 A video introducing single-cell RNA sequencing of spider embryos was uploaded to YouTube.
21.02.20 A video showing a lecture Fujiwara gave to the general people at BRH was uploaded to YouTube. "Mathematics and Physics for Biological Shaping."
21.02.17 Ryota Nanjo presented his master thesis work in Osaka Univ. Title is "Comparison of Chromatin Dynamics in Arthropod Embryogenesis: Analysis of Parasteatoda tepidariorum early embryos by ATAC sequencing."
21.01.19 Mainichi shinbun picked up our long-term activities using spiders.
20.12.07 Another news paper (Asahi shinbun) picked up our research activity.
20.11.12 A news paper (Mainichi shinbun) picked up our research activity.
20.11.07 Yasuko and Hiroki were invited to participate in Public Journal Club Developmental Biology online.
20.10.19 Our articles on the model spider Parasteatoda appeared in a Japanese scientific journal.
20.10.17  A video showing our system for spider egg collection was uploaded to YouTube.
A video showing our system for spider egg collection was uploaded to YouTube.
20.09.11 Hiroki gave a talk in an online symposium "Protostome functional genetics ‐ Tribolium and friends"
20.09.10 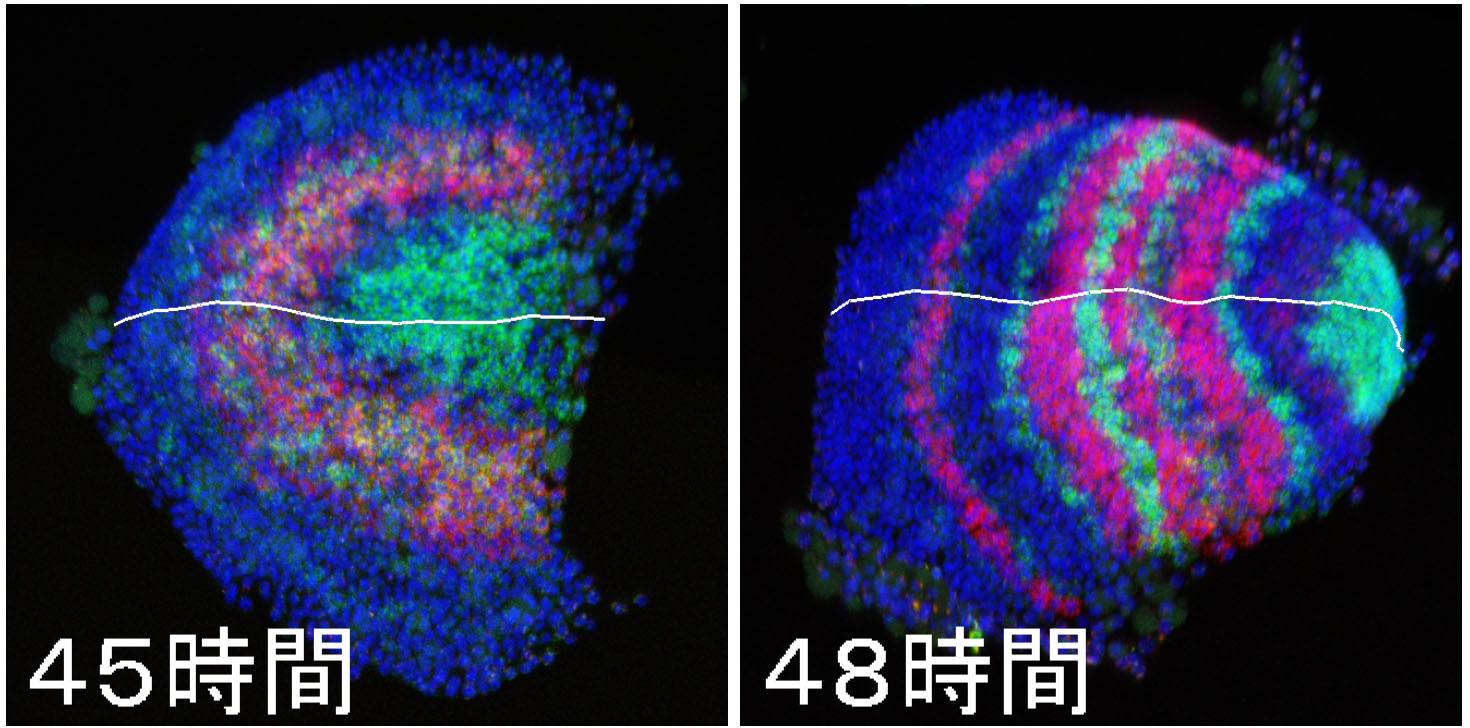 Our paper on spider segmentation appeared in Science Advances. For many years, Yasuko made tremendous efforts to complete it.
Our paper on spider segmentation appeared in Science Advances. For many years, Yasuko made tremendous efforts to complete it.
20.06.20 A video showing our everyday research life was uploaded to YouTube.
20.06.01 Asahi shinbun picked up our cadherin study.
20.05.16 Hiroki gave a lecture on a surprising spider world using a video.
20.04.07 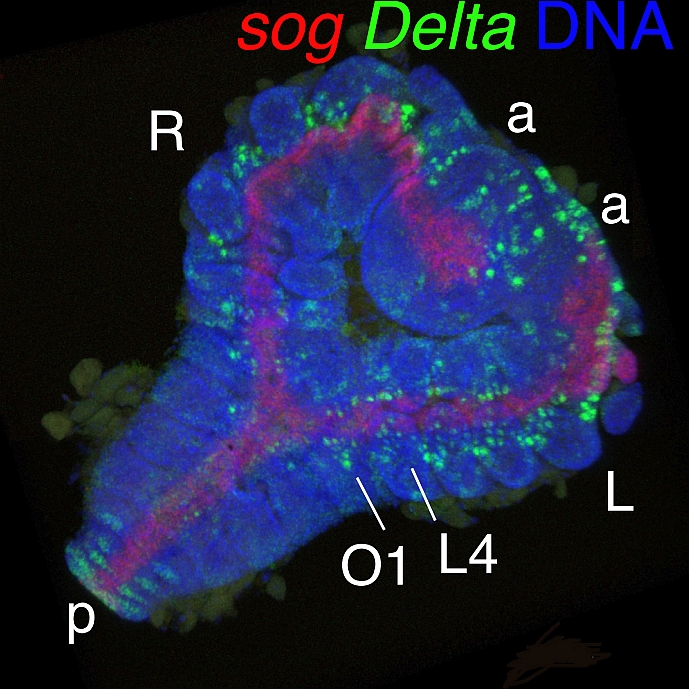 Our paper on the regulative nature of spider embryos appeared in the spider issue of Development Genes and Evolution.
Our paper on the regulative nature of spider embryos appeared in the spider issue of Development Genes and Evolution.
20.03.23 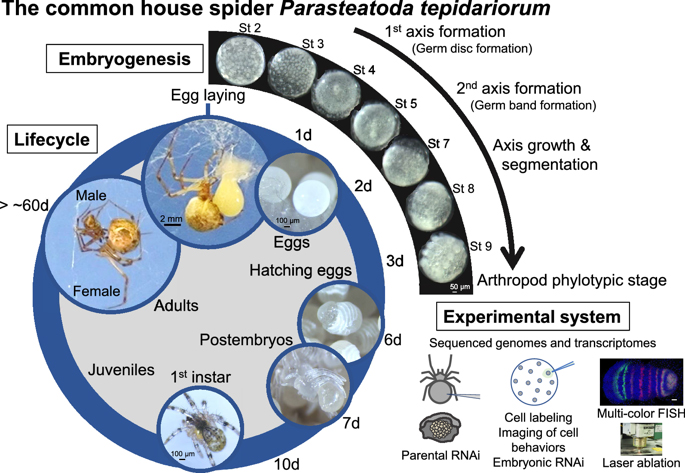 Our review article on the common house spider appeared in EvoDevo.
Our review article on the common house spider appeared in EvoDevo.
20.01.03 Our DATA article appeared in Data in Brief.
Research content
Multicellular animals, the Metazoa, show a great diversity of body forms. Fossil records suggest that the major morphology-based taxonomic groups of metazoans, including the arthropods and chordates, had already existed more than 500 million years ago. Comparative studies of genetic mechanisms underlying the development of body forms in the model vertebrates and invertebrates, such as Xenopus and Drosophila, have revealed some conserved aspects of the developmental mechanisms among the metazoans. However, our understanding of the deeply ancient animals from which the body-form diversity was evolved is still in its infancy. For example, there is no consensus of idea about whether the segmented body plans in the major metazoan groups have common or independent origins. One of the biggest obstacles to reconstructing the macroevolutionary processes is the fact that during evolution the animals flexibly and sometimes drastically changed their genetic programs without disrupting the basic body plan in each major metazoan branch. Our research in BRH aims to overcome this problem and achieve a good understanding of the ancestral animals that were allowed to evolve the great diversity of animal forms. To this end, we compare mechanisms of development and cell-cell adhesion mainly using Drosophila and the spider Parasteatoda tepidariorum (see below for more details).
.jpg)
▲A fiy and spider in amber
▼Members
▼Latest publication New!
▼Award New!
▼Activity Reports New!
▼Diary New!
▼Our research projects
▼Recent topics in our laboratory
▼Videos
▼Databases New!
MemberMore
Publication List
[Research] Nanoscale Visualization of Drosophila E-cadherin Ectodomain Interactions Using DNA Origami Nanoblocks
In this study, we applied DNA origami technology to reconstitute and visualize DE-cadherin ectodomain adhesive interactions. Read our paper.
[Research] Discovery of a lineage-specific, fast-evolving gene that regulates early spider development
We found fuchi nashi, a lineage-specific, fast-evolving GATA-like gene that contributes to zygotic activation of genes involved in endoderm specification and pattern formation in the early spider embryo. Read our paper.
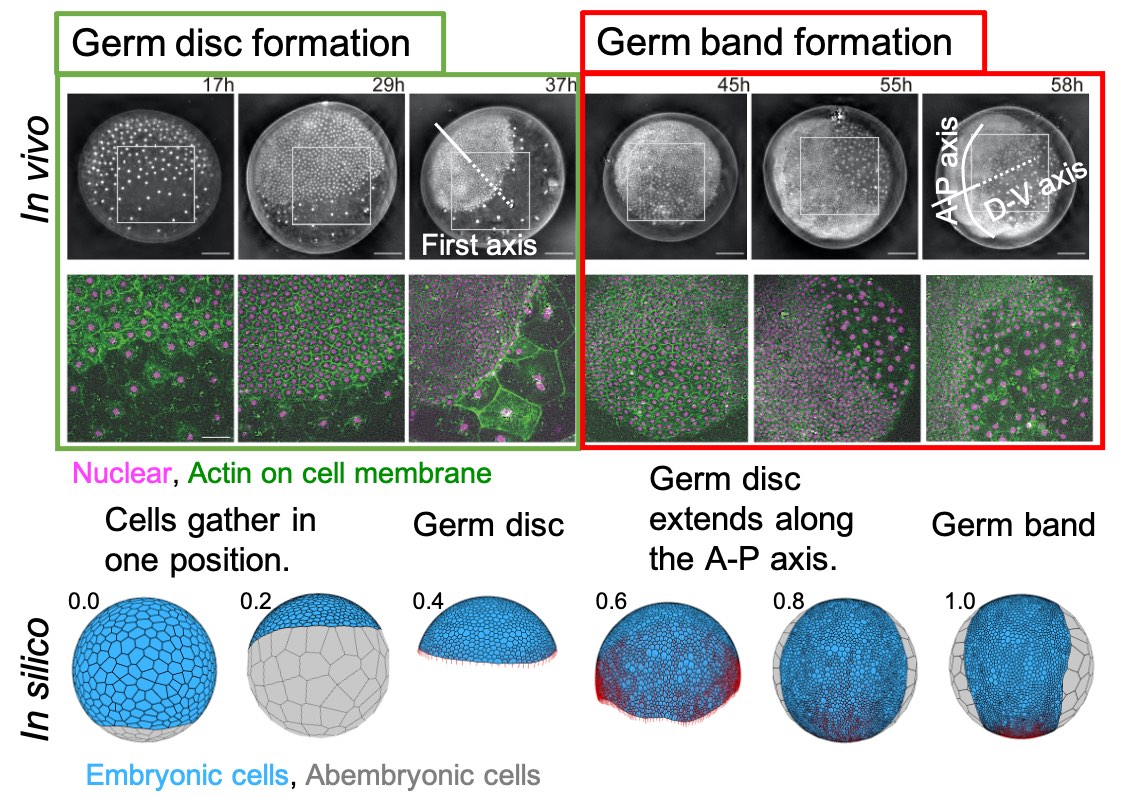
[Research] Virtual cells shape an arthropod embryo on the computer
Our mathematical model mimicking embryonic cell dynamics can reproduce the spider embryo shaping in silico. Read our paper.
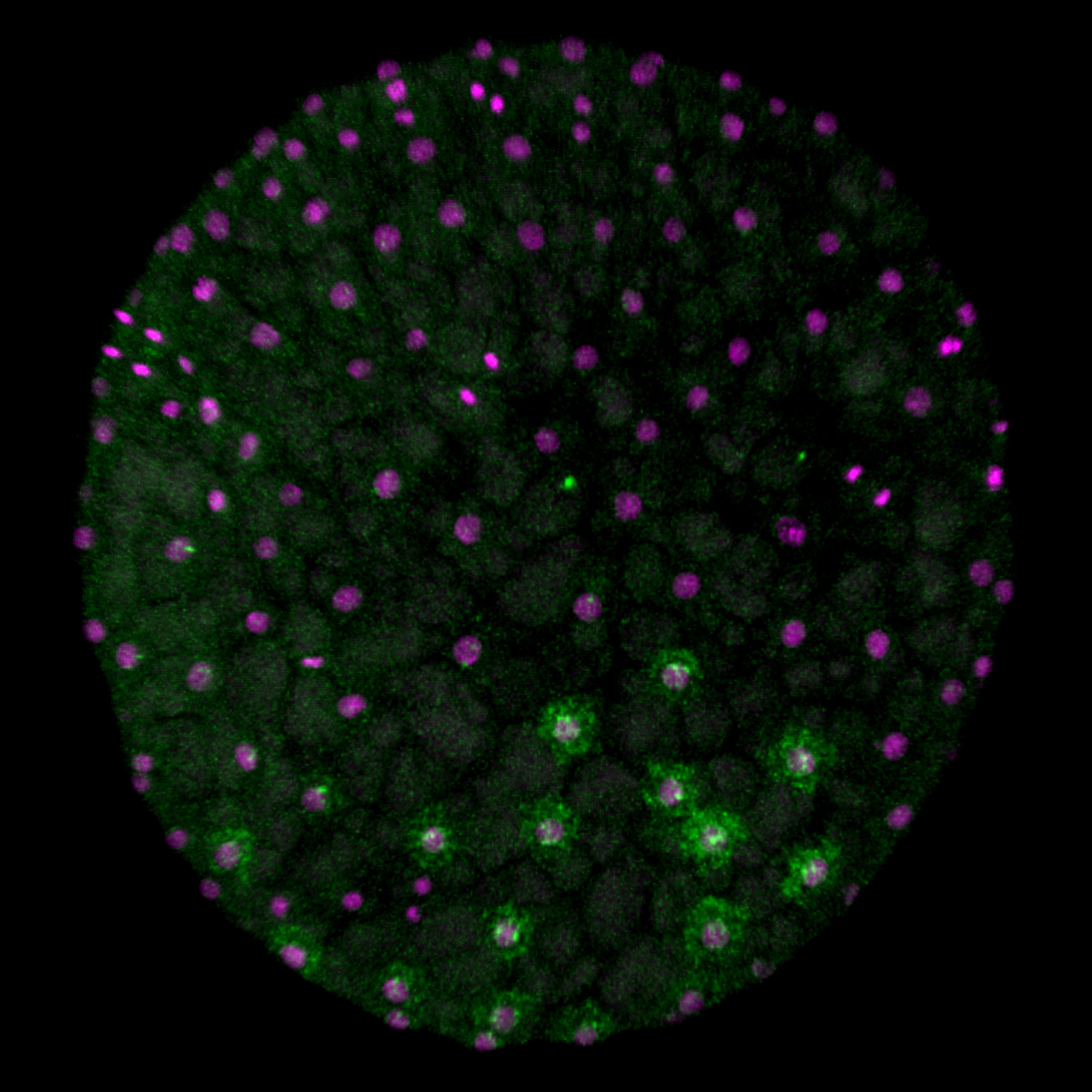
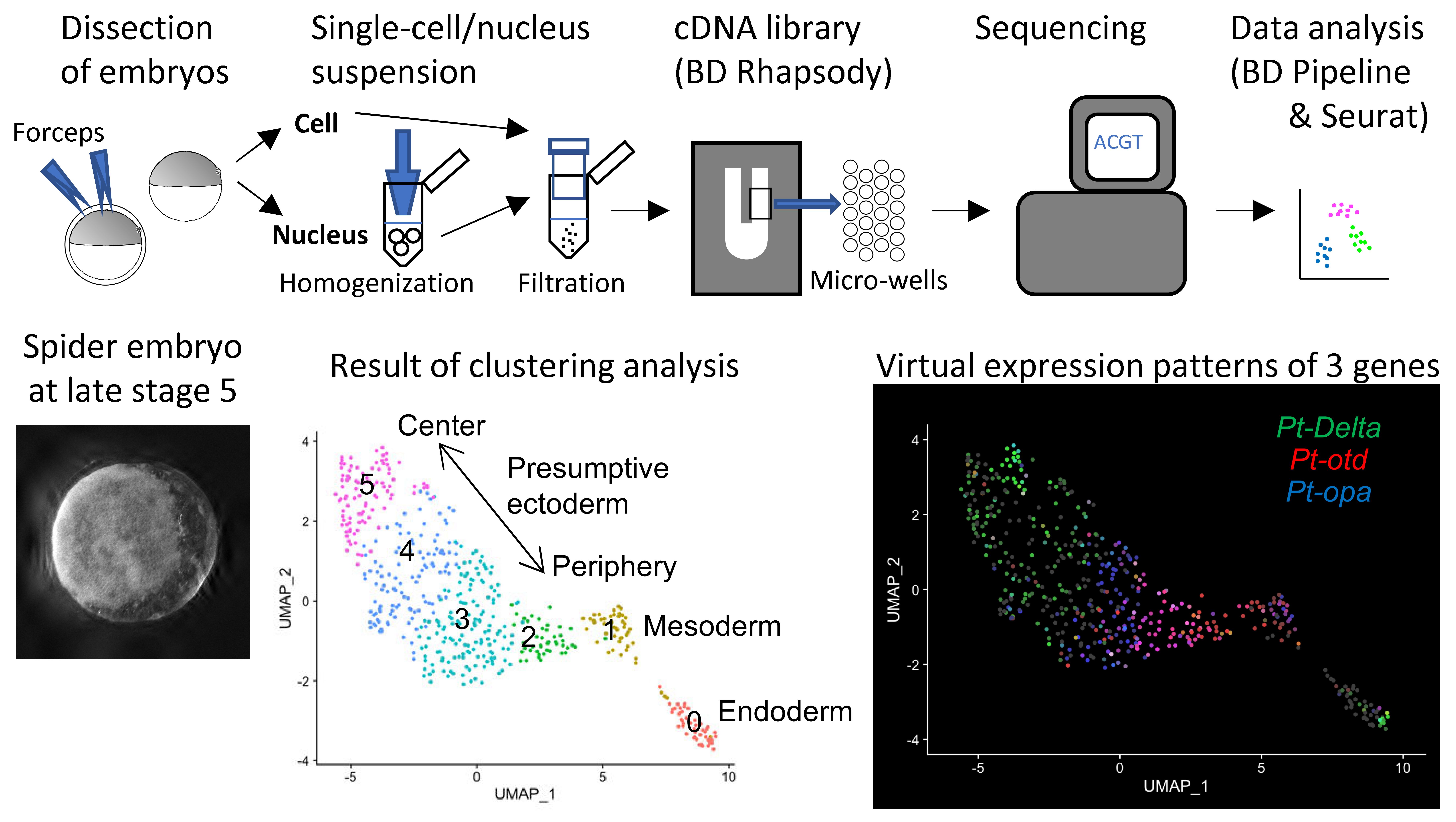
[Research] Reconstruction of the axis of a spider embryo on the computer after cell dissociation
In this work, we applied single-cell and single-nucleus RNA sequencing to early embryos in the common house spider Parasteatoda tepidariorum. These techniques require cell dissociation prior to obtaining transcriptomic data, which leads to a loss of information about the position of individual cells in the embryo. It is usually difficult to overcome this demerit, but our experiments using spider embryos allowed us to reconstruct a correct axis of the spider embryo on the computer. Read our paper.
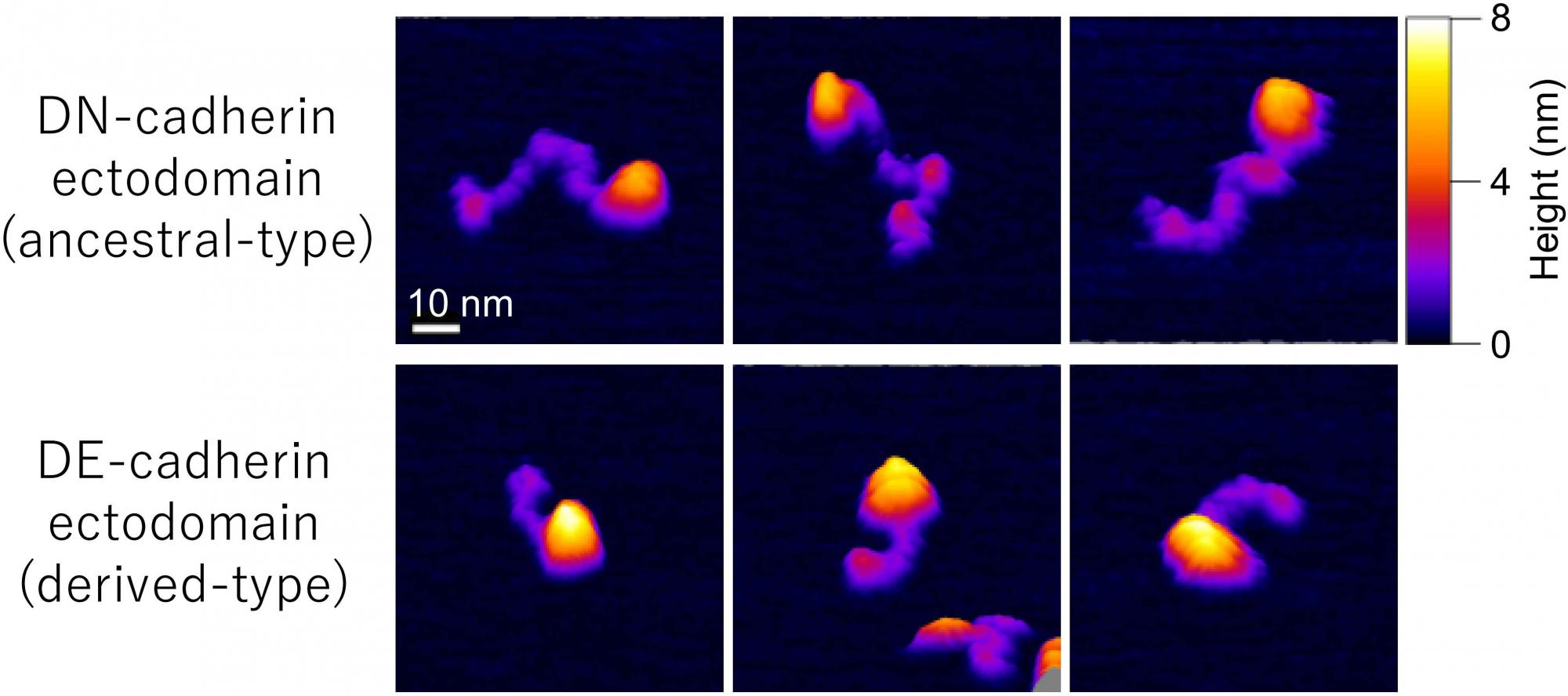
[Research] Visualization of ancestral long cadherin by atomic force microscopy
Classical cadherins constitute a family of Ca2+-dependent cell-cell adhesion molecules that contribute to the construction and maintenance of the multi-celllular animal body. Despite the fundamental importance of this molecular family, they exhibit a great diversity in the domain composition and organization of the ectodmain depending on the phylogeny. The fruit fly Drsoophila melanogaster has two classical cadherins, one of which is ancestral, known as DN-cadherin, the other is derived, known as DE-cadherin. These two cadherins provide good examples with which we can investiagte the strucutral and functional relationships between the ancestral and derived states of classical cadherin. In this work, we successfully purified DN- and DE-cadherin ectodomains and visualized their molecular morphologies using high-speed atomic force microscopy(HS-AFM). Read more.
[Research] Dynamics and diversity in spider segmentation
We genome-widely identified 98 genes which transcriptionally responded to altered states of Hedgehog signaling. Our analyses focused on the msx1 gene, which is negatively regulated by Hedgehog signaling. We found that this msx1 is a key segmentation gene in the spider embryo. Its expression behaves as waves and its function is requied to generate repetitive units in the embryonic field. Read more.
[Research] Our mini-review on the common house spider appears in a renowned academic journal
We have published a mini-review in the international academic journal EvoDevo that introduces the common house spider Parasteatoda tepidariorum as an emerging experimental system. You can know the characteristics of this spider species, what kind of research themes can be addressed, and what experimental techniques can be used with this spider. The sequence of the genome of P. tepidariorum has been read and published, and rich transcriptomic resources are available. As spiders of this species live close to us, we believe that you can easily start using them not only for research purposes but also for education purposes. If interested, please read this mini-review. Our databases site also may be useful for getting information on this spider (https://www.brh2.jp).
Hiroki Oda and Yasuko Akiyama-Oda
The common house spider Parasteatoda tepidariorum
EvoDevo (2020) 11:6
https://doi.org/10.1186/s13227-020-00152-z
[Research] Duplication of body axes experimentally induced in spider embryos
We have published a review paper that describes twinning of spider embryos experimentally induced as well as provides a historical background on such twinning experiments in spiders. One of the conclusions of this review paper is "The two concepts of developmental biology, organizer and self-regulation, which were described in vertebrate embryology, are similarly applied in the development of spiders." Please see two movies showing body axes duplications following two different types of embryological manipulations (Movie1 and Movie2), as well as a movie showing delayed body axes formation following removal of cumulus by laser irradiation (Movie3).
Hiroki Oda, Sawa Iwasaki-Yokozawa, Toshiya Usui, and Yasuko Akiyama-Oda
Experimental duplication of bilaterian body axes in spider embryos: Holm’s organizer and self-regulation of embryonic fields
Development Genes and Evolution, 10 April 2019, First Online (Open Access)
DOI: 10.1007/s00427-019-00631-x
https://link.springer.com/article/10.1007/s00427-019-00631-x
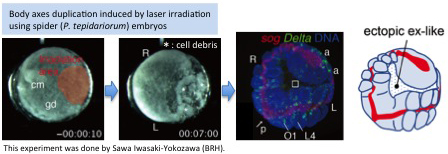
■Movie1
Body axes duplication by transplantation of cumulus using jumping spider (Hasarius adansoni) embryos
Find more details.
https://link.springer.com/article/10.1007/s00427-019-00631-x
■Movie2
Body axes duplication by laser irradiation using house spider (Parasteatoda tepidariorum) embryos
Find more details.
https://link.springer.com/article/10.1007/s00427-019-00631-x
■Movie3
Removal of cumulus by laser irradiation using house spider (Parasteatoda tepidariorum) embryos
Find more details.
https://link.springer.com/article/10.1007/s00427-019-00631-x
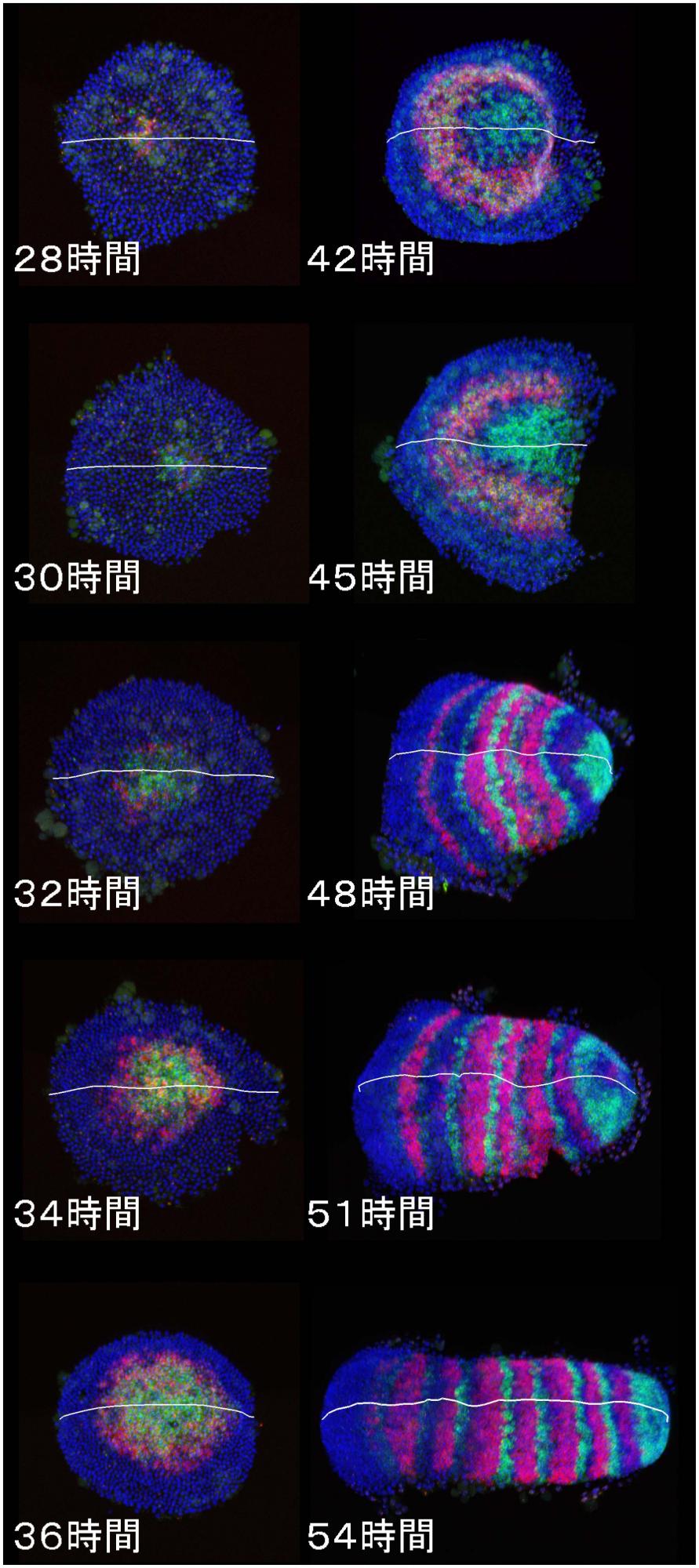
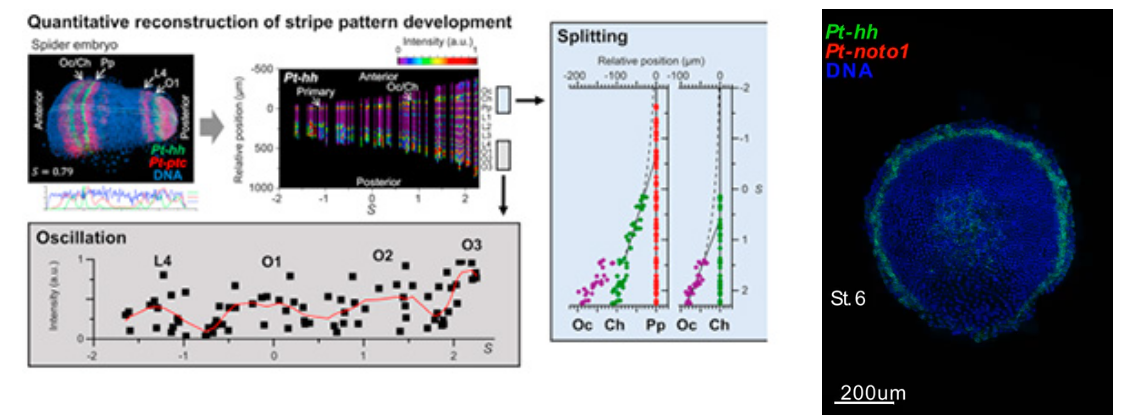
- [Research] Diversity of stripe-forming processes in an embryo
Our paper, which describes a quantitative study of stripe-forming processes relevant to segmentation in a spider embryo, has been published in the journal Developmental Biology. Segmented body patterns along the body axis are shared morphological characters of arthropods. Experimental studies using various insect and other arthropod models, however, have suggested that developmental processes resulting in striped patterns considerably vary depending on the body region as well as on the species. But, such varied stripe-forming processes have not been documented in a common framework. In this work, using embryos of the spider Parasteatoda tepidariorum, we quantitatively characterized the diversity of stripe-forming processes in a dynamic cellular field. Cell behaviors were also examined to show wave-like features of gene expression. Our descriptions highlight contrasting pattern dynamics, splitting versus oscillation, in both terminal (head and abdominal) regions of the field. In the middle (thoracic) region, stripes appear to form by a third mechanism. Employment of a diversity of segmentation processes in an embryonic field, which are region-dependent, is common among arthropods.
Natsuki Hemmi, Yasuko Akiyama-Oda, Koichi Fujimoto, and Hiroki Oda(2018)
A quantitative study of the diversity of stripe-forming processes in an arthropod cell-based field undergoing axis formation and growth
Developmental Biology 437 (2) :84-104
DOI: 10.1016/j.ydbio.2018.03.001
https://www.sciencedirect.com/science/article/pii/S0012160617309089
- [Research]
Schwager, Sharma, Clarke, Leite, Wierschin, Pechmann, Akiyama-Oda, Esposito, Bechsgaard, Bilde, Buffry, Chao, Dinh, Doddapaneni, Dugan, Eibner, Extavour, Funch, Garb, Gonzalez, Gonzalez, Griffiths-Jones, Han, Hayashi, Hilbrant, Hughes, Janssen, Lee, Maeso, Murali, Muzny, da Fonseca, Paese, Qu, Ronshaugen, Schomburg, Schönauer, Stollewerk, Torres-Oliva, Turetzek, Vanthournout, Werren, Wolff, Worley, Bucher, Gibbs, Coddington, Oda, Stanke, Ayoub, Prpic, Flot, Posnien, Richards and McGregor(2017)
The house spider genome reveals an ancient whole-genome duplication during arachnid evolution
BMC Biology, Volume 15, Issue 62, 2017
https://bmcbiol.biomedcentral.com/articles/10.1186/s12915-017-0399-x

- [Research]
Mizuki Sasaki, Yasuko Akiyama-Oda, and Hiroki Oda(2017)
Evolutionary origin of type IV classical cadherins in arthropods
BMC Evolutionary Biology, Volume 17, Issue 142, June
DOI: 10.1186/s12862-017-0991-2
https://bmcevolbiol.biomedcentral.com/articles/10.1186/s12862-017-0991-2
- [Research]
Shigetaka Nishiguchi, Akira Yagi, Nobuaki Sakai, Hiroki Oda(2016)
Divergence of structural strategies for homophilic E-cadherin binding among bilaterians
Journal of Cell Science DOI: 10.1242/jcs.189258
http://jcs.biologists.org/content/129/17/3309
Award
Our paper was selected for DB Outstanding Paper Award 2018
The journal Developmental Biology announced that they selected our paper for DB Outstanding Paper Award 2018 (see the journal site ). The first author Natsuki Hemmi will receive the award. She is invited to give a talk in the SDB 78th annual meeting held in Boston this July.
Click here for the contents of Natsuki's invited presentation at the SDB meeting.
Natsuki Hemmi, Yasuko Akiyama-Oda, Koichi Fujimoto, and Hiroki Oda(2018)A quantitative study of the diversity of stripe-forming processes in an arthropod cell-based field undergoing axis formation and growth Developmental Biology 437 (2) :84-104. DOI: 10.1016/j.ydbio.2018.03.001
https://www.sciencedirect.com/science/article/pii/S0012160617309089

■Congratulations, Natsuki! Great talk in Boston
Hemmi Natsuki, M.S. (alumnus of our laboratory) was invited to the 78th Annual Meeting of Society for Developmental Biology held in Boston from July 26th to 29th, 2019 and gave a talk about the research that was awarded the DB Outstanding Paper Award 2018 (click here for details:). I felt how much the audience was moving during her talk. I am sure that it had been really hard for her to prepare the presentation while working for a company and was proud that she made a great presentation in English. At the banquet on the last day, the prize award ceremony was held. Many participants talked to her "Congratulations!", “The videos were amazing” , and so on. I also made a poster presentation. I used the full presentation time (from 20:00 to 23:00) to explain our spider study. Although most of the people there were unfamiliar with research using spider embryos, they looked interested in our spider work. It was an awesome night. Even late in the night, there were a lot of participants discussing their work with loud voice. I felt the power of America. (Akiyama)
Click here for the contents of Natsuki's presentation at the SDB meeting in Boston.

Activity Reports
BRH welcomed Dr. Eric Wieschaus and Dr. Trudi Schüpbach, who were interested in seeing spider embryos
On May 14, 2019, Dr. Eric F. Wieschaus, Squibb Professor in Molecular Biology at Princeton University and 1995 Nobel laureate, and Dr. Trudi Schüpbach, Emeritus Professor of Molecular Biology at Princeton University, visited us the Biohistory Research Hall (BRH) before attending the Annual Meeting of the JSDB held in Osaka.(Read more→)

Diary
[April 2019]
Sharing and effective use of our BRH data resources
Nearly two years have passed since we started to run our BRH data resources, site where you can search several kinds of databases…[read more]
[April 2013]
Our Collaborator Maarten Hilbran,Ph.D. who is from Oxford Brookes University
stayed our laboratory for 2 weeks.
Our research projects
1)Evolution of adherens junction cadherins
2)Development of the spider embryo
1) Evolution of classical cadherins responsible for adherens junctions
We investigate the diversification of the structure and function of cell-cell adhesion molecules called classical cadherins, which are the key components of the adherens junctions. This junction type is of particular importance in driving and regulating tissue morphogenesis. We have identified lineage-specific structural reductions in the extracellular regions of classical cadherins. Our data suggest that similar but independent structural changes occurred in the classical cadherins at the adherens junctions in several different animal lineages. We are now attempting to investigate the functional significance of the classical cadherin structural changes using Drosophila. We want to understand the impact of evolutionary changes in cell-cell adhesion interfaces on metazoan morphogenesis.
2) Development of the spider embryo
Second, using the spider Parasteatoda tepidariorum, a common house spider, we try to identify cellular, genetic and molecular mechanisms of arthropod body plan formation that are distinct from those of Drosophila. We have found that Hedgehog signaling plays key roles in anterior-posterior axis formation in the spider (Fig. 2). In Drosophila, similar roles are played by the Drosophila Bicoid-Nanos system, which is operational only in the syncytial environment. We also showed that cellularization in the spider is complete at or before the 16-nucleus stage, sharply contrasting with the situation of the Drosophila embryo, which is a syncytium until the number of nuclei increases to 6,000. Our findings have revealed that the common arthropod body plan is achieved by largely distinct mechanisms, suggesting that genetic programs underlying the formation of the basic body plan significantly changed during evolution. One of the most important messages from our spider studies is that the diversity of the genetic programs is large even within each major metazoan subbranch (such as the arthropod phylum). Comparative studies of widely-ranged animal species within the subbranch are required to better understand the ancestral states of the arthropod body plan and other bilaterally symmetrical metazoan body plans.


An adult female of the spider Parasteatoda tepidariorum (left) and a developing embryo (right)
Recent topics in our laboratory
1) A "domain-loss" model for classical cadherin evolution
We proposed a “domain-loss” model to explain the diversification of the extracellular structures of classical cadherins that are responsible for adherens junction assembly in the metazoans.
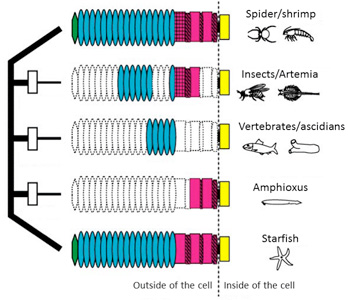
Fig. 1. A “domain-loss” model to explain the diversification of classical cadherin structure.
See more details
Oda and Takeichi, 2011
Oda et al., 2005
2) A shortened cadherin can function in adhesion, but not in epithelial bending
We constructed a Drosophila DE-cadherin lacking a half of its extracellular region and found that it can function as an adhesion molecule to organize the blastoderm epithelium but not as a morphogenetic regulator to drive ventral furrow formation, an epithelial bending accompanied by cooperative apical constrictions of cells.
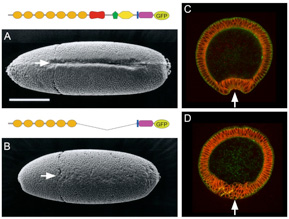
Fig.2. Replacement of DE-cadherin with a shortened one caused specific defects in ventral furrow formation.
See more details
Oda and Takeichi, 2011
Haruta et al., 2010
3) Hedgehog signaling in global pattering of the sider embryo
We discovered key roles of Hedgehog signaling in anterior-posterior axis formation in the early embryo of the spider Parasteatoda tepidariorum. We also showed that migration of the source of Dpp signal that orients the dorsal-ventral axis depends on the roles of Hedgehog signaling. This work highlighted the crucial importance of Hedgehog signaling in global patterning of the spider embryo.
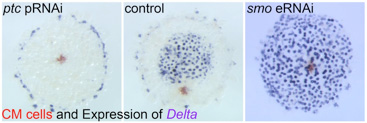
Fig. 3. Embryonic RNA interference for Hedgehog signaling pathway components results in severe defects in early embryonic patterning.
4) Microinjection is now available for early spider embryos
We succeeded in applying a microinjection technique to early spider embryos. Using this technique, we can perform, for example, cell labeling, exogenous expression of fluorescent protein and embryonic RNA interference. Actually, we used this technique to verify that cellularization is complete at or before the 16-nucleus stage in the spider embryo.
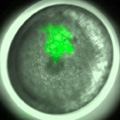
Fig. 4. Cell clone labeling by microinjection of FITC-dextran into single blastomeres in early spider embryos.
5) Stripe-splitting, a novel mode of animal segmentation
We discovered a novel mode of segmentation, stripe-splitting, in the developing head of the spider Parasteatoda tepidariorum embryo. A stripe of hedgehog expression, after traveling, undergoes repeated splitting to pattern the three segmental units in the spider head. Our data suggest that this segmentation strategy employs an autoregulatory signaling network that hedgehog signaling pathway components, orthodenticle and odd-pared participate in. Our findings provide new insights into the evolution of segmented body plans in the metazoans.
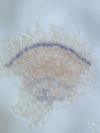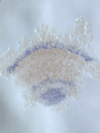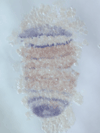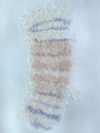
Fig. 5. A stripe of hedgehog expression (purple) in the head region (see upper than the brown region) undergoes repeated splitting.